BIRCH
Workflow example: Use BLASTP
to search for sequences related to a query protein and retrieve hits
The pea defense gene DRR206 confers strong resistance to fungal
pathogens when transformed into Brassica
napus. Although no Brassica
homologue of this gene has been found by hybridization, we would like
to see if a homologous sequence can be found in speices closely-related
to B. napus. Since protein
searches are more sensitive than DNA
searches, we need to get the amino acid sequence from the genomic
sequence.
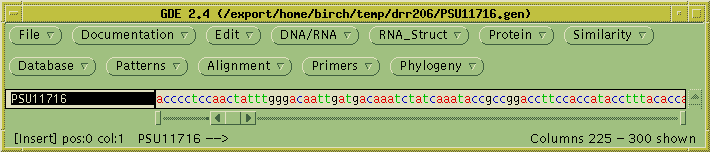
The first step is to read in GenBank file for the genomic sequence.
Save the file PSU1716l.gen in your
current working directory. In
GDE, choose File --> Open,
and read in the file.
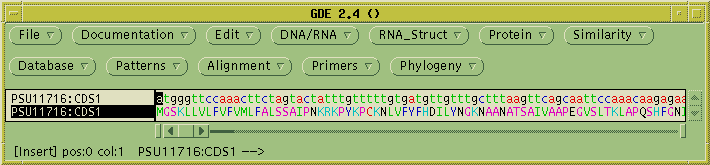
Next, we need to extract the protein coding sequence from the genomic
sequence. In GenBank files, coding sequences are annotated as CDS
features. Choose Database -->
Features (extract by feature keys):

Set the feature key to CDS, Database to "Selected sequences", and send
output to GDE. A new GDE window pops up with the coding sequence which
was extracted from the larger genomic sequence. To translate into
protein, select PSU11716:CDS1, and choose DNA/RNA --> Ribosome.
Where the menu says "Send output to a new GDE window", choose "No." The
translated protein will be sent to the current GDE window.
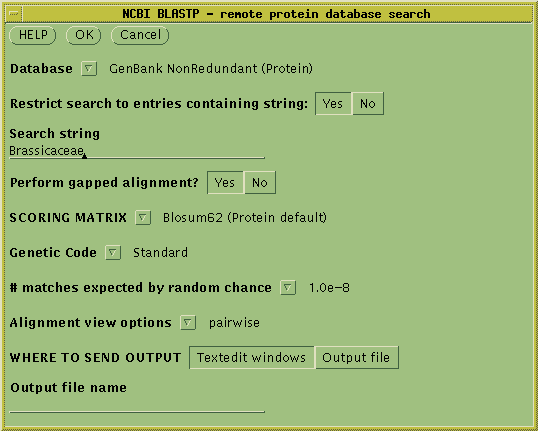
To run the BLASTP search, select the amino acid sequence and choose
Database --> NCBI BLASTP.
Choose the GenBank NonRedundant (Protein)
database. To limit the search to relatives of Brassica napus, click
"Yes" for Restrict search to entries containing string:, and type
"Brassicaceae" as the search string. To eliminate poor matches, set the
# matches expected by random chance to 1.0e-8 (ie. 10-8).
Click on OK to start the search.
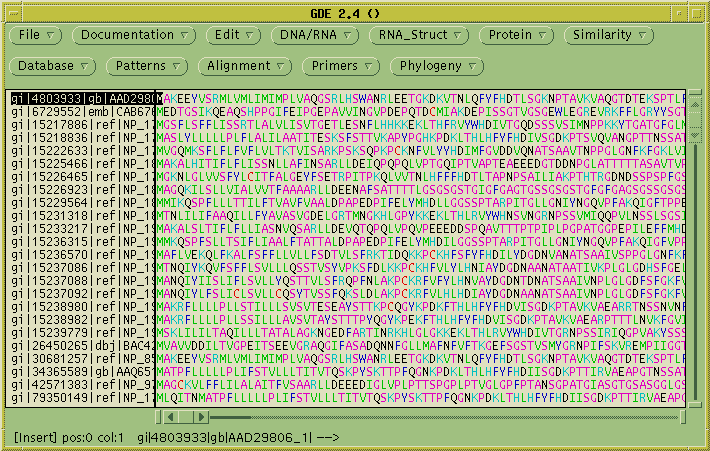
The query sequence is sent to the NCBI Blast server, and the results
are returned when complete. The results appear in several windows. The
Blast report appears in a text editor
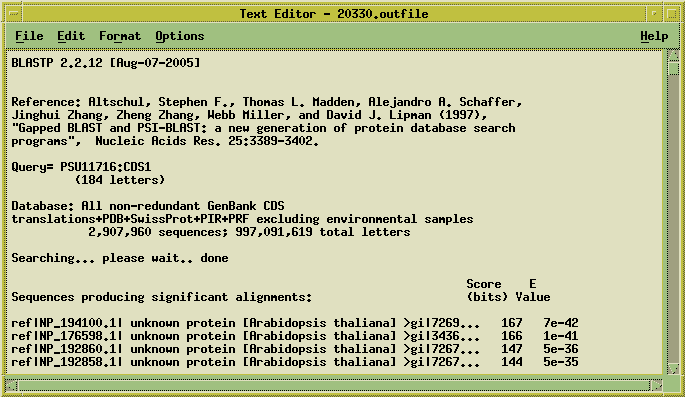
and the Accession numbers of the hits are sent to another text editor
window:
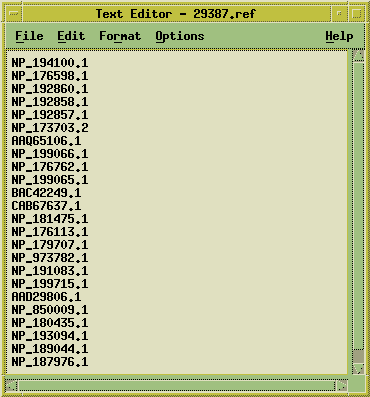
The Accession numbers are also sent to a dGDE window. dGDE is a program
for working with lists of numbers, such as accession numbers, TaxID
numbers etc. dGDE is designed to send requests to the SeqHound data
warehouse (http://www.blueprint.org/seqhound),
and retrieve information from SeqHound. Since SeqHound can
only retrieve sequences using GI numbers, we first need to convert the
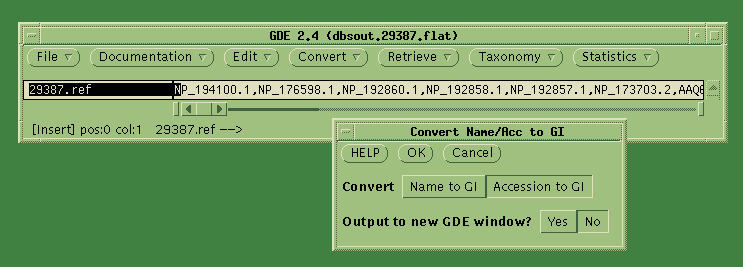
Accession numbers into GI numbers. Choose Convert --> Convert
Name/Acc to GI. Click on "Accession to GI", and send the output
to the
current GDE window (ie. No).
The Accession numbers are sent to SeqHound, and the GI numbers are read
into the GDE window. Select the GI numbers, and choose Retrieve -->
Protein from Protein GI. Click OK.
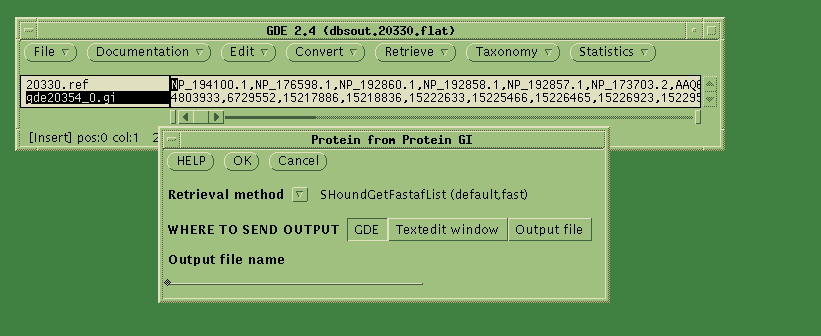
GDE will send the GI numbers to SeqHound, calling the SeqHound method
SHoundGetFastaList. The sequences are returned to a new GDE window: